Mac OS X R from CRAN. On Mac OS X if the only version of R you have installed is the standard R distribution from CRAN then RStudio will by default run against the.
Peter Langfelder and Steve Horvath
with help of many other contributors
Semel Institute for Neuroscience and Human Behavior, UC Los Angeles (PL),
Contains R 3.4.4 framework, R.app GUI 1.70 in 64-bit for Intel Macs, Tcl/Tk 8.6.6 X11 libraries and Texinfo 5.2. The latter two components are optional and can be ommitted when choosing 'custom install', they are only needed if you want to use the tcltk R package or build package documentation from sources. To find out your R version, look at the message you get whenever R starts up, or run sessionInfo. If you are running a newer version of R than 3.4.4, I would not recommend downgrading unless you’re sure you know what you’re doing! That said, older R binaries are available on CRAN. There are different pages for different platforms. R 3.4.0 Download Mac Usb Audio Codec Driver Download Mac How To Download Redis On Mac Winrar For Mac free. download full Gmod Prop Hunt Download For Mac Waves Plugin Bundle Mac Free Download Download Free Ftp Client For Mac Numark Cue 7 Download For Mac Citrix Receiver 11.5 Mac Download.
Dept. of Human Genetics and Dept. of Biostatistics, UC Los Angeles (SH)Peter (dot) Langfelder (at) gmail (dot) com,SHorvath (at) mednet (dot) ucla (dot) edu
BMC Bioinformatics, 2008 9:559(link opens in a new tab/window)
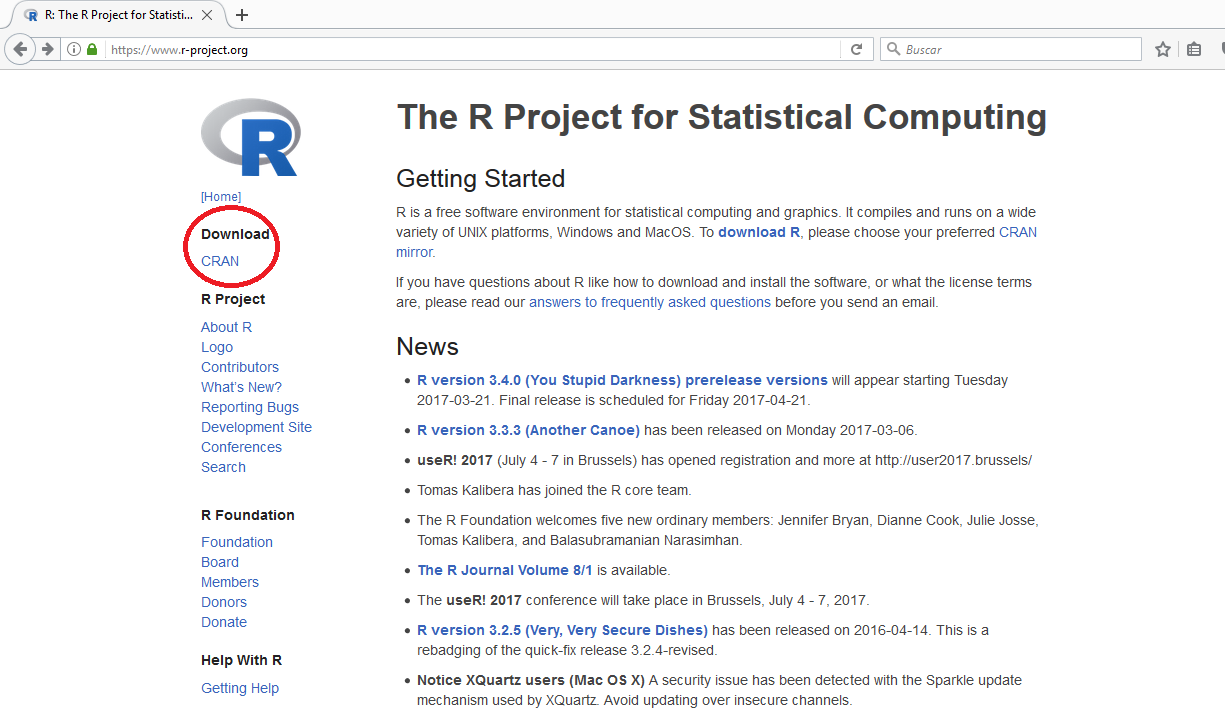
Quick navigation
Abstract
Correlation networks are increasingly being used inbioinformatics applications. For example,weighted gene co-expression network analysis is a systems biologymethod for describing the correlation patterns among genesacross microarray samples. Weighted correlation network analysis(WGCNA) can be used for finding clusters (modules) of highly correlatedgenes, for summarizing such clusters using the moduleeigengene or an intramodular hub gene, for relating modules to one another and to external sample traits(using eigengene network methodology), and for calculating modulemembership measures. Correlation networks facilitate network basedgene screening methods that can be used to identify candidatebiomarkers or therapeutic targets. These methods have beensuccessfully applied in various biological contexts, e.g. cancer,mouse genetics, yeast genetics, and analysis of brain imaging data. While parts ofthe correlation network methodology have been described in separate publications, there is aneed to provide a user-friendly, comprehensive, and consistentsoftware implementation and an accompanying tutorial.The WGCNA R software package is a comprehensive collection of Rfunctions for performing various aspects of weighted correlationnetwork analysis. The package includes functions for networkconstruction, module detection, gene selection, calculations oftopological properties, data simulation, visualization, andinterfacing with external software. Along with the R package wealso present R software tutorials. While the methods developmentwas motivated by gene expression data, the underlying data miningapproach can be applied to a variety of different settings.

Getting started with R and Weighted Gene Co-expression Network Analysis
The package described here is an add-on for the statistical language and environment R (freesoftware).Our tutorials contain step bystep instructions such that even complete novice users should be able to get started in R immediately.Readers wishing to learn about the theory and published applications of WGCNA are invited tovisit the WGCNAmain page.
R Tutorials
A comprehensive set of tutorials that illustrate various aspects of WGCNA is available. We offer not only introductory tutorials thatintroduce basic functionality of the package, but also more advanced analyses in which we used the WGCNApackage in our own research.Click here to access the tutorial page.
Further reading
R 3.4 For Mac High Sierra
Peter Langfelder occasionally writes about WGCNA features and other topicsrelating to data analysis. The articles are written for a general audience and try to avoid deep technical details. We also have a few technical reports that discuss aselected deeply technical aspects of the WGCNA methodology - these are more mathematical and targeted primarilly todie-hard statistician geeks.Automatic installation from CRAN
The WGCNA package is now available from the Comprehensive R Archive Network (CRAN), the standard repositoryfor R add-on packages. Currently, some of the required packages is only available from Bioconductor and needto be installed using Bioconductor's installation tools. The easiest way to do this is
BiocManager::install('WGCNA')
The first command (install.packages('BiocManager')) can be skipped if the package BiocManager is already installed.
This will install the WGCNA package and all necessary dependencies. The catch is that this only installsthe newest version of WGCNA if your R version is also the newest (minor) version.Users using older versions of R will need to follow the manual download andinstallation instructions below.
Note for Mac users: CRAN occasionally fails to compile the WGCNA package for Mac OS X. This leads to the error message 'Package WGCNA is not available..' when callingBiocManager::install(). If this occurs, please download the binary version from here and follow the installationinstructions (or, if you are able to compile packages locally, download the source and install that).
Note of caution: The newest versions of WGCNA is available from CRAN only for the current Rversion and (usually) one older version. For example, if your R version is 3.2.1 and the current R version on CRAN is 3.5.0, the automaticinstallation and update will not use the newest version of WGCNA. Please update your R to the newest versionor use the manual download below.

Problems installing or using the package? Please see our list of frequently askedquestions. Your problem and the solution may already be posted there.
Manual download and installation
Please follow these steps only if the automatic package installation above does not work.
Prerequisites:
The current version of the WGCNA package will only work with R version 3.0.0 and higher. If youhave an older version of R, please upgrade your R.
The WGCNA package requires the following packages to be installed: stats, grDevices, utils, matrixStats (0.8.1 or higher), Hmisc, splines, foreach, doParallel, fastcluster, dynamicTreeCut,survival, parallel, preprocessCore, GO.db, impute, and AnnotationDbi.If your system does not have them installed, the easiestway to install them is to issue the following command at the R prompt:
source('http://bioconductor.org/biocLite.R')
biocLite(c('GO.db', 'preprocessCore', 'impute'))
R 3.4 For Mac Catalina
Please note that GO enrichment calculations in WGCNA are deprecated; we recommend using the R package anRichment which provides replacement for WGCNA functions GOenrichmentAnalysis() anduserListEnrichment().
If you run an older version of R, the above may not install the newest version of the dynamicTreeCutpackage. Should you encounter this problem, please manually download and install dynamicTreeCut from this web page.
R package download and installation:Package WGCNA_1.69-81 (last updated 2020/04/30) is available here as source code and several pre-compiledversions for various platforms. In general it is preferable to download the source and compile the packagelocally; however, if this is not practical, please select an appropriate compiled version.
- Source for Linux and all users able to compile the package locally: WGCNA_1.69-81.tar.gz
- Compiled binary Mac OS X:WGCNA_1.69.tgz
- Comiled binary for Windows running R-3.4.0 or higher: WGCNA_1.69.zip
- Reference manual in pdf format
- Quick reference: overview table of most important functions
- A terse changelog
The package version numbers follow the formatpackageName_major.minor-revision. Minor versions typically add or change some functionality;revisions typically contain bugfixes or minor enhancements.
Should you discover bugs (of which there are most likely plenty), please report them to Peter Langfelder.
Problems installing or using the package
Please see our list of Frequently Asked Questions (and frequently given answers);the solution to your problem may lie there. In particular, you can find answers about spurious Macerrors, compatibility problems when upgrading WGCNA, and others. If you still cannot solve the problem, email PeterLangfelder.Old versions of R package WGCNA
Older version of the packages presented on this page are available here.Citing the WGCNA package
If you use WGCNA in published work, please cite it to properly credit people who have created it.Can i cancel a tool box order for mac tools windows 10. The WGCNA as an analysis method is described in
- Zhang B and Horvath S (2005) A General Framework for Weighted Gene Co-Expression Network Analysis, Statistical Applications in Genetics and Molecular Biology: Vol. 4: No. 1, Article 17 PMID: 16646834
The package implementation is described in the article
If you use any q-value (FDR) calculations, please also cite at least one of the following articles:
- Storey JD. (2002) A direct approach to false discovery rates. Journal of the Royal Statistical Society, Series B, 64: 479-498.
- Storey JD and Tibshirani R. (2003) Statistical significance for genome-wide experiments. Proceedings of the National Academy of Sciences, 100: 9440-9445.
- Storey JD, Taylor JE, and Siegmund D. (2004) Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: A unified approach. Journal of the Royal Statistical Society, Series B, 66: 187-205.
If you use the collapseRows function to summarize/convert probe-level data to gene-leveldata, please cite
R 3.4 For Mac Shortcut
- Miller JA, Cai C, Langfelder P, Geschwind DH, Kurian SM, Salomon DR, Horvath S (2011) Strategies foraggregating gene expression data: The collapseRows R function. BMC Bioinformatics 12:322.
If you use module preservation calculations, please cite
- Langfelder P, Luo R, Oldham MC, Horvath S (2011) Is my network module preserved and reproducible? PloSComp Biol. 7(1): e1001057
If you use functions rgcolors.func, plotCor, plotMat,stat.bwss, or stat.diag.da, please also cite the article
- Sandrine Dudoit, Yee Hwa Yang, Matthew J. Callow, and Terence P. Speed, Statistical methods for identifying differentially expressed genes in replicated cDNAmicroarray experiments, STATISTICA SINICA 2002, 12:111
Acknowledgments
The core of the functions and other code was written by Peter Langfelder and Steve Horvath, partlybased on older code written by Steve Horvath and Bin Zhang. Multiple people contributed additional code,most prominently Jeremy Miller, Chaochao (Ricky) Cai, Lin Song, Jun Dong, and Andy Yip. The package alsocontains code adapted from external packages that were either orphaned (such as package
sma) ortheir development has made the code difficult to use in WGCNA (such as package qvalue). A big thanks goes out to people who continue report the many bugs in the package. The package is currently maintained by Peter Langfelder.
This directory contains binaries for a base distribution and packages to run on Mac OS X (release 10.6 and above). Mac OS 8.6 to 9.2 (and Mac OS X 10.1) are no longer supported but you can find the last supported release of R for these systems (which is R 1.7.1) here. Releases for old Mac OS X systems (through Mac OS X 10.5) and PowerPC Macs can be found in the old directory.
Note: CRAN does not have Mac OS X systems and cannot check these binaries for viruses.Although we take precautions when assembling binaries, please use the normal precautions with downloaded executables.
Package binaries for R versions older than 3.2.0 are only available from the CRAN archive so users of such versions should adjust the CRAN mirror setting (https://cran-archive.r-project.org) accordingly. Ubuntu 32 bit iso.
R 4.0.3 'Bunny-Wunnies Freak Out' released on 2020/10/10
Please check the MD5 checksum of the downloaded image to ensure that it has not been tampered with or corrupted during the mirroring process. For example type
openssl sha1 R-4.0.3.pkg
in the Terminal application to print the SHA1 checksum for the R-4.0.3.pkg image. On Mac OS X 10.7 and later you can also validate the signature using
pkgutil --check-signature R-4.0.3.pkg
Latest release:
| R-4.0.3.pkg (notarized and signed) SHA1-hash: 8402f586aef1fdb12c6e34c73b286f87318fb1be (ca. 85MB) | R 4.0.3 binary for macOS 10.13 (High Sierra) and higher, signed and notarized package. Contains R 4.0.3 framework, R.app GUI 1.73 in 64-bit for Intel Macs, Tcl/Tk 8.6.6 X11 libraries and Texinfo 6.7. The latter two components are optional and can be ommitted when choosing 'custom install', they are only needed if you want to use the tcltk R package or build package documentation from sources. Note: the use of X11 (including tcltk) requires XQuartz to be installed since it is no longer part of OS X. Always re-install XQuartz when upgrading your macOS to a new major version. Important: this release uses Xcode 10.1 and GNU Fortran 8.2. If you wish to compile R packages from sources, you will need to download and GNU Fortran 8.2 - see the tools directory. |
| NEWS (for Mac GUI) | News features and changes in the R.app Mac GUI |
| Mac-GUI-1.73.tar.gz SHA1-hash: 7f4b1d050757ce78545bdeb9d178a69d13046aa1 | Sources for the R.app GUI 1.73 for Mac OS X. This file is only needed if you want to join the development of the GUI, it is not intended for regular users. Read the INSTALL file for further instructions. |
Note: Previous R versions for El Capitan can be found in the el-capitan/base directory.Binaries for legacy OS X systems: | |
| R-3.6.3.nn.pkg (signed) SHA1-hash: c462c9b1f9b45d778f05b8d9aa25a9123b3557c4 (ca. 77MB) | R 3.6.3 binary for OS X 10.11 (El Capitan) and higher, signed package. Contains R 3.6.3 framework, R.app GUI 1.70 in 64-bit for Intel Macs, Tcl/Tk 8.6.6 X11 libraries and Texinfo 5.2. The latter two components are optional and can be ommitted when choosing 'custom install', they are only needed if you want to use the tcltk R package or build package documentation from sources. |
| R-3.3.3.pkg MD5-hash: 893ba010f303e666e19f86e4800f1fbf SHA1-hash: 5ae71b000b15805f95f38c08c45972d51ce3d027 (ca. 71MB) | R 3.3.3 binary for Mac OS X 10.9 (Mavericks) and higher, signed package. Contains R 3.3.3 framework, R.app GUI 1.69 in 64-bit for Intel Macs, Tcl/Tk 8.6.0 X11 libraries and Texinfo 5.2. The latter two components are optional and can be ommitted when choosing 'custom install', it is only needed if you want to use the tcltk R package or build package documentation from sources. Note: the use of X11 (including tcltk) requires XQuartz to be installed since it is no longer part of OS X. Always re-install XQuartz when upgrading your OS X to a new major version. |
| R-3.2.1-snowleopard.pkg MD5-hash: 58fe9d01314d9cb75ff80ccfb914fd65 SHA1-hash: be6e91db12bac22a324f0cb51c7efa9063ece0d0 (ca. 68MB) | R 3.2.1 legacy binary for Mac OS X 10.6 (Snow Leopard) - 10.8 (Mountain Lion), signed package. Contains R 3.2.1 framework, R.app GUI 1.66 in 64-bit for Intel Macs. This package contains the R framework, 64-bit GUI (R.app), Tcl/Tk 8.6.0 X11 libraries and Texinfop 5.2. GNU Fortran is NOT included (needed if you want to compile packages from sources that contain FORTRAN code) please see the tools directory. NOTE: the binary support for OS X before Mavericks is being phased out, we do not expect further releases! |
Subdirectories:
| tools | Additional tools necessary for building R for Mac OS X: Universal GNU Fortran compiler for Mac OS X (see R for Mac tools page for details). |
| base | Binaries of R builds for macOS 10.13 or higher (High Sierra) |
| contrib | Binaries of package builds for macOS 10.13 or higher (High Sierra) |
| el-capitan | Binaries of package builds for OS X 10.11 or higher (El Capitan build) |
| mavericks | Binaries of package builds for Mac OS X 10.9 or higher (Mavericks build) |
| old | Previously released R versions for Mac OS X |
You may also want to read the R FAQ and R for Mac OS X FAQ. For discussion of Mac-related topics and reporting Mac-specific bugs, please use the R-SIG-Mac mailing list.
Information, tools and most recent daily builds of the R GUI, R-patched and R-devel can be found at http://mac.R-project.org/. Please visit that page especially during beta stages to help us test the Mac OS X binaries before final release!
Package maintainers should visit CRAN check summary page to see whether their package is compatible with the current build of R for Mac OS X.
Binary libraries for dependencies not present here are available from http://mac.R-project.org/libs and corresponding sources at http://mac.R-project.org/src.
Last modified: 2020/10/10, by Simon Urbanek
